This post accompanies our new paper in Wellcome Open Research.
Regular readers of this blog will know that I worry about uncertainty in the numbers we are using to model drug action on electrophysiology quite a lot – see our recent white paper for an intro where we discuss the various sources of uncertainty in our simulations, and a 2013 paper on variability observed when you repeat ion channel screening experiments lots of times. That paper studied variability in the averages of lots of experiments, in contrast, it is also possible to look at the uncertainty that remains when you just do one experiment (or one set of experiments).
We have been looking at screening data that were published recently by Crumb et al. (2016). This is an exciting dataset because it covers an unprecedented number of ion currents (7), for a good number of compounds (30 – a fair number anyway!). The ideal scenario for the CiPA initative is that we can feed these data into a mathematical model, and classify compounds as high/intermediate/low arrhythmic risk as a result. Of course, the model simulation results are only ever going to be as accurate as the data going in, I’ve sketched what I mean by this in the plot below.

Figure 1: Uncertainty Quantification. Here we imagine we have two inputs (red, blue) to a nonlinear model, and we observe the resulting model outputs. Top: for just a single value (with no associated uncertainty). Middle: for two well-characterised (low uncertainty) inputs, these give us distinct probabilistic model predictions. Bottom: for two uncertain and overlapping inputs, inevitably giving us uncertain and overlapping output distributions.
In Figure 1 we can see the same scenario viewed three different ways. We have two inputs into the simulation – a ‘red’ one and a ‘blue’ one. You could think about these as any numbers, e.g. “% block of an ion current”.
Top row of Fig 1
If we ignored uncertainty, we might do what I’ve shown on the top row: plug in single values; and get out single outputs. Note that blue was higher than red and gives a correspondingly higher model output/prediction. But how certain were we that red and blue took those values? How certain are we that the output really is higher for blue?
Middle row of Fig 1
Instead of just the most likely values, it is helpful to think of probability distributions for red and blue, as I’ve shown in the middle row of Figure 1. Here, we aren’t quite sure of their exact values, but we are fairly certain that they don’t overlap (this step of working out distributions on inputs is called “uncertainty characterisation“), so their most likely values are the same as the ones we used before in the top row. When we map these through our model* (a step called “uncertainty propagation“) we get two probability distributions. N.B. these output distributions are not necessarily the same shape as the ones that went in, for a nonlinear model. In the sketch the output distributions don’t overlap much either. But this is only guaranteed to be true if the model output is a linear (with non-zero slope!) function of one input; in general, the output distributions could easily overlap more than the inputs [e.g. imagine the model output is a sine wave function of the input with red input = blue input + 2π]; which is why uncertainty propagation is an important exercise! The whole process of uncertainty characterisation and propagation is known as uncertainty quantification.
Bottom row of Fig 1
In the bottom row of Figure 1 we see another scenario – here there is lots of overlap between the red and blue input distributions. So we think blue is higher than red, but it could easily be the other way around. Now we map these through our model, and get correspondingly large and overlapping probability distributions on our outputs. Even in the best-case scenario (i.e. a linear model output depending solely on this input), we would end up with the same distribution overlap as the inputs; and for non-linear models, where outputs are complicated functions of many inputs, the distributions could overlap much more. So bear in mind that it isn’t possible to get less-overlapping distributions out of the model than the ones that went in, but it is possible to get more overlapping ones. The uncertainty always increases if anything (I hadn’t really thought about it before, but that might be related to why we think about entropy in information theory?).
If we consider now that the output is something like ‘safety risk prediction for a drug’, could we distinguish whether red or blue is a more dangerous drug? Well, maybe we’ll be OK with something like the picture in the middle row of Figure 1. Here my imaginary risk indicator model output distinguishes between the red and blue compounds quite well. But we can state categorically “no” if the inputs overlap to the extent that they do in the bottom row, before we even feed them in to a simulation. So we thought it would be important to do uncertainty characterisation and work out our uncertainty in ion channel block that we have from dose-response curves; before doing action potential simulations. This is the focus of our recent paper in Wellcome Open Research**.
Uncertainty Characterisation
In the new paper, Ross has developed a Bayesian inference method and code for inferring probability distributions for pIC50 values and Hill coefficients.
The basic idea is shown in Figure 2, where we compare the approach of fitting a single vs. distribution of dose-response curves:

Figure 2: best fit versus inference of dose-response curves. On the right we have plotted lots of samples from the distribution of possible curves, with each sample equally likely, so the darker regions are the more likely regions. Data from Crumb et al. (2016).
On the left of Figure 2 we see the usual approach that’s taken in the literature, fitting a line of best fit through all the data points. On the right, we plot samples of dose-response curves that may have given rise to these measurements.
The method works by inferring the pIC50 and Hill coefficients that describe the curve, but also the observation error that is present in the experiment. i.e the ‘statistical model’ is:
data = curve(pIC50, Hill) + Normal noise (mean = 0, standard deviation = σ).
One of the nice features of using a statistical model like this is that it tries to learn how much noise is on the data by looking at how noisy the data are, and therefore generates dose-response curves spread appropriately for this dataset, rather than with a fixed number for the standard deviation of the observational noise.
At this point we calculate what the % block of the current (inputs into model) might look like from these inferred curves, this is shown in Figure 3:

Figure 3: uncertainty characterisation. When we go to do a simulation we’ll want the “% hERG block” as an input into the simulations. This figure shows how we work that out at a given concentration. Imagine drawing a line vertically in the top plot (same as right plot in Fig 2), now we just look at the distribution along this line, which is displayed in the bottom plot for two concentrations (green and pink-ish).
The new paper also extends this approach to hierarchical fitting (saying data from each experiment were generated with different pIC50 and Hill coefficients), but I will let you read the paper for more information on that.
So what can we conclude about the distribution of inputs for the Crumb dataset? Well, that’s a little bit tricky since it’s 7 channels and therefore a seven dimensional thing to examine. To have a look at it we took 500 samples of each of the seven ion current % blocks. This gives a table where each row looks like:
Drug (1-30) | Sample (1-500) | % block current 1 | % block current 2 | ... | % block current 7 |
So the seven % blocks are the seven axes or dimensions of our input dataset.
I then simply did a principal components analysis that will separate out the data points as much as possible by projecting the data onto new axes that are linear combinations of the old ones. You can easily visualize up to 3D, as shown in the video below.
In the video above you see the samples for each compound plotted in a different colour (the PCA wasn’t told about the different compounds). So each cloud is in a position that is determined by what % block of each channel the compounds produce at their free Cmax concentrations (given in the Crumb paper). What we see is that about 10 to 12 of the compounds are in distinct distributions, so they block currents in a combination unlike other compounds, i.e. behave like the different inputs in the middle row of Figure 1. But the remaining ones seem to block in distributions that could easily overlap, like the inputs in the bottom row of Figure 1. As you might expect, this is clustered close to the origin (no block) co-ordinate.
Here, these first three principal components happen to describe 94% of the variation in the full 7D dataset, so whilst it is possible that there is some more discrimination between the ‘clouds’ of samples for each compound in higher dimensions, your first guess would be that this is not likely (but this is a big caveat that needs exploring futher before reaching firm conclusions). So it isn’t looking promising that there is enough information here, in the way we’ve used it anyway, to distinguish between the input values for half of these compounds.
Uncertainty Propagation
But once you’ve got this collection of inputs you can simply do the uncertainty propagation step anyway and see what comes out. We did this for a simple O’Hara 1Hz steady state pacing experiment, applying conductance block according to the video’s cloud values for each of the 30 compounds, to work out predicted output distributions of APD90 for each compound. The results are shown in Figure 4.
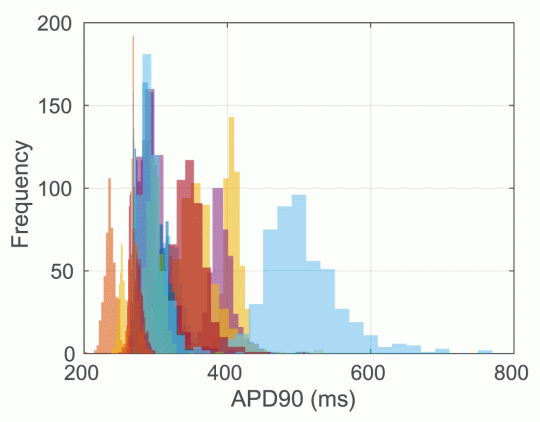
Figure 4: distributions of simulated APD90 in the O’Hara model (endocardial variant) at free Cmax for all 30 compounds in the Crumb dataset, using samples from the inferred dose-response curves (Figure 15 in the paper). Random colours for each of the 30 compounds.
Figure 4 is in line with what we expected from the video above, so it causes us to have some pause for thought. Perhaps five compounds have distinctly ‘long’ APD, and perhaps two have distinctly ‘short’ APD, but this leaves 23 compounds with very much overlapping ‘nothing-or-slight prolongation’ distributions. The arrhythmic risk associated with each of these compounds is not entirely clear (to me!) at present, so it is possible that we could distinguish some of them, but it is looking a bit as if we are in the scenario shown in the bottom right of Figure 1 – and this output overlaps to such an extent that it’s going to be difficult to say much.
So we’re left with a few options:
- Do more experiments (the larger the number of data points, the smaller the uncertainty in the dose-response curves), whether narrower distributions allow us to classify the compounds according to risk remains to be seen.
- Examine whether we actually need all seven ion currents as inputs (perhaps some are adding noise rather than useful information on arrhythmic risk).
- Get more input data that might help to distinguish between compounds – the number one candidate here would be data on the kinetics of drug binding to hERG.
- Examine other possible outputs (not just APD) in the hope they distinguish drugs more, but in light of the close-ness of the input distributions, this is perhaps unlikely.
So to sum up, it is really useful to do the uncertainty characterisation step to check that you aren’t about to attempt to do something impossible or extremely fragile. I think we’ve done it ‘right’ for the Crumb dataset, and it suggests that we will need more, or less(!), or different data to distinguish pro-arrhythmic risk. Comments welcome below…
Footnotes:
* The simplest way to do this is to take a random sample of the input probability distribution, run a simulation with that value to get an output sample, then repeat lots and lots of times to build up a distribution of outputs. This is what’s known as a Monte Carlo method (using random sampling to do something that in principle is deterministic!). There are some cleverer ways of doing it faster in certain cases, but we didn’t use them here!
** By the way: Wellcome Open Research is a new open-access, open-data, journal for Wellcome Trust funded researchers to publish any of their findings rapidly. It is a pre-print, post-publication review journal, so reviewers are invited after the manuscript is published and asked to help the authors revise a new version. The whole review process is online and open, and it follows the f1000research.com model. So something I’m happy we’ve supported by being in the first issue with this work.

In the crumb paper I noticed they used point estimates for free cmax as the drug concentration. So they assume that there is no uncertainty in plasma protein binding, which there clearly is, especially when you get > ~96% bound. At that point no-one knows what is truly “free”. Also you have huge variations in drug concentrations from patient to patient. Then you add on top of that the uncertainty in the model parameters. I think you can guess where I am going with this…
Makes you think about what Figure 4 might actually look like. I guess it’s easy to start with quantifying uncertainty in an assay that’s routinely used versus uncertainty in the mean free cmax etc. Makes you wonder which process has the largest influence on the width of the histograms in Figure 4.
Yep – good point, the Cmax certainly has uncertainty and maybe systematic bias in heart-muscle concentration too. If it could be quantified, then this probabilisitic framework could take it into account too (along with uncertainty in model parameters, structure, etc. etc.). This is very much the start of the process rather than the end! We’ve been looking at how the variability in measurements like this relates to how well we make rabbit wedge predicitons at GSK, so we’re starting to get an impression of the relative sizes of various sources of uncertainty. Future post on that early in the new year I hope!
This is a really interesting paper and thanks for the blog post to explain it further. In the cancer community there is a lot of interest in generating dose response data for anti-cancer compounds in a panel of cancer cell lines, and then building models using genetic features (mutations, gene expression etc) to explain differential sensitivity. I wonder whether your approach could help this process by allowing the uncertaintly in the original dose response measurement to be carried through to the modelling? See:
http://www.nature.com/nchembio/journal/v12/n2/full/nchembio.1986.html
http://www.sciencedirect.com/science/article/pii/S0092867416307462
Glad you liked it, thanks for the feedback. I think the approach would work for any dose-response stuff, and it certaintly makes sense to see how robust any subsequent predictions are to this kind of uncertainty in model inputs. So yes, I think it’s a good idea!
Practically, we might need to modify the code if extra parameters such as saturation-level of the dose-response curve were required (the drug/ion-channel stuff tends to saturate at 100%, so this number is hard-coded without any uncertainty in it), but that would be do-able.